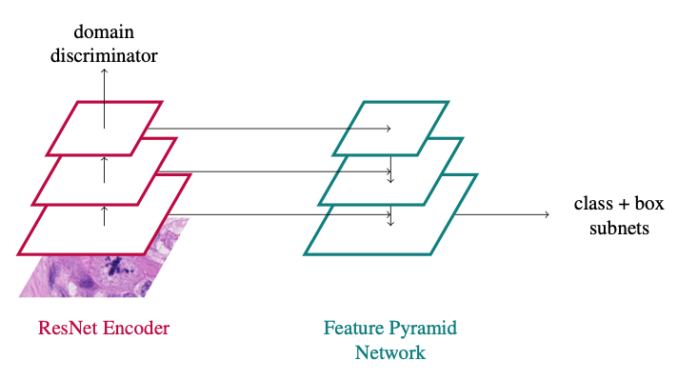
We provided a reference docker container to all participants that was done by Frauke Wilm (FAU, Germany). Since the approach seems harder to beat than we expected, we prepared a two-pager on arxiv.org to let you know how exactly she did it. Of course the approach was not optimized on the test set or the preliminary test set in any way.
In principle, the approach is a vanilla RetinaNet approach with a domain adversarial branch added:
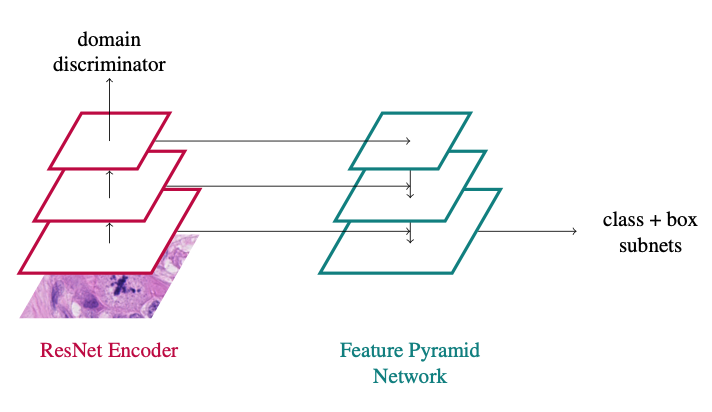
You can find all the details in the paper:
Wilm, F., Breininger, K., Aubreville, M: Domain Adversarial RetinaNet as a Reference Algorithm for the MItosis DOmain Generalization (MIDOG) Challenge, https://arxiv.org/abs/2108.11269

Hello Marc,
thank you very much for this great challenge. I’m just wondering why you are releasing Frauke Wilm’s reference method before the deadline. For me, and I bet for some others as well, it’s actually pretty easy to add this method to an existing pipeline (i.e. I’ve already implemented it in my networks, but not tested it). Using this feature right now would feel like cheating. So is the purpose of this blog post to make use of your experience, or what was the intent?
Dear Jakob,
there’s a number of reasons for that:
Have fun in the challenge! Best,
Marc
Okay, thanks for the fast reply.
It seems that the best method in the leaderboard achieves F1-score of 0.7514. exactly equal to this baseline algorithm ?
Yes. But submitting the baseline does not count.
Hi, according to preprint report, I think this baseline approach didn’t utilize the hard negative label. Am I right ?
Yes, that is correct. We experimented with both but ultimately decided to only train with the mitotic figure labels.